Kubeflow’s superfood for Data Scientists
TL;DR: Kale lets you deploy Jupyter Notebooks that run on your laptop or on the cloud to Kubeflow Pipelines, without requiring any of the Kubeflow SDK boilerplate. You can define pipelines just by annotating Notebook’s code cells and clicking a deployment button in the Jupyter UI. Kale will take care of converting the Notebook to a valid Kubeflow Pipelines deployment, taking care of resolving data dependencies and managing the pipeline’s lifecycle.
Introduction
Kale was introduced to the Kubeflow community during a community call on May 14th (recording) as a tool to seamlessly deploy annotated Jupyter Notebooks to Kubeflow Pipelines. The idea was to allow ML practitioners to focus on their code, while still being able to make use of Kubeflow Pipelines.
Since then, we have worked hard on this project to provide a tool that can really streamline day-to-day MLOps and lower the barrier to entry of the Kubeflow ecosystem. First off, Kale has now its own GitHub organization (github.com/kubeflow-kale), where you can find the main Kale repository with the backend implementation, a dedicated repository for the brand new JupyterLab UI (more on that later in the post) and a curated set of examples to showcase the power of Kale.
In this blog post we are going to reiterate on the motivations that led us to build Kale, how Kubeflow can benefit from this project, the major features of the recent updates and some helpful resources to show how easy it is to setup Kale and start playing with the Notebooks.
Motivation
Data science is inherently a pipeline workflow, from data preparation, to training and deployment, every ML project is organized in these logical steps. Without enforcing a strict pipeline structure to Data Science projects, it is often too easy to create messy code, with complicated data and code dependencies and hard to reproduce results. Kubeflow Pipelines is an excellent tool to drive data scientists to adopt a disciplined (“pipelined”) mind set when developing ML code and scaling it up in the Cloud.
The Kubeflow Pipelines’ Python SDK is a great tool to automate the creation of these pipelines, especially when dealing with complex workflows and production environments. Still, when presenting this technology to ML researchers or Data Scientists that don’t have strong software engineering expertise, KFP can be perceived as too complex and hard to use. Data Science is often a matter of prototyping new ideas, exploring new data and models, experimenting fast and iteratively. In these scenarios one would prefer to just run some rough code and analyze the results rather than setting up complex workflows with a specific SDK.
In our experience, this experimentation phase often happens directly in Jupyter Notebooks, where a Data Scientist exploits the power of interactivity and visualizations of Jupyter. Then, refactoring a messy Notebook to an organized KFP pipeline can be a challenging and time-consuming task.
Kale
Kale was designed to address these difficulties by providing a tool to simplify the deployment process of a Jupyter Notebook into Kubeflow Pipelines workflows. Translating Jupyter Notebook directly into a KFP pipeline ensures that all the processing building blocks are well organized and independent from each other, while also leveraging on the experiment tracking and workflows organization provided out-of-the-box by Kubeflow.
Fun Fact:
When Kale was becoming more that just a prototype, we wondered what name we could give to this new project. We wanted it to resonate with Jupyter, so we looked into the Jupyter’s (planet) moons and satellites. Kale (pronounced /ˈkeɪliː/), one of the small and unheard of Jupyter’s moon, became our top choice thanks to the fitting acronym. During an after hours work day, the logo came into existence after discovering (to our utter bewilderment) that Kale was actually a vegetable as well! Obviously many people started calling Kale with its (correct) vegetable pronunciation, instead of our initially intended Jupyter moon name. So we resigned to just having a “super food” for data scientists, and leave any connection to planets and greek deities in our memories.
The main idea behind Kale is to exploit the JSON structure of Notebooks to annotate them, both at the Notebook level (Notebook metadata) and at the single Cell level (Cell metadata). This annotations allow to:
- Assign code cells to specific pipeline components
- Merge together multiple cells into a single pipeline component
- Define the (execution) dependencies between them
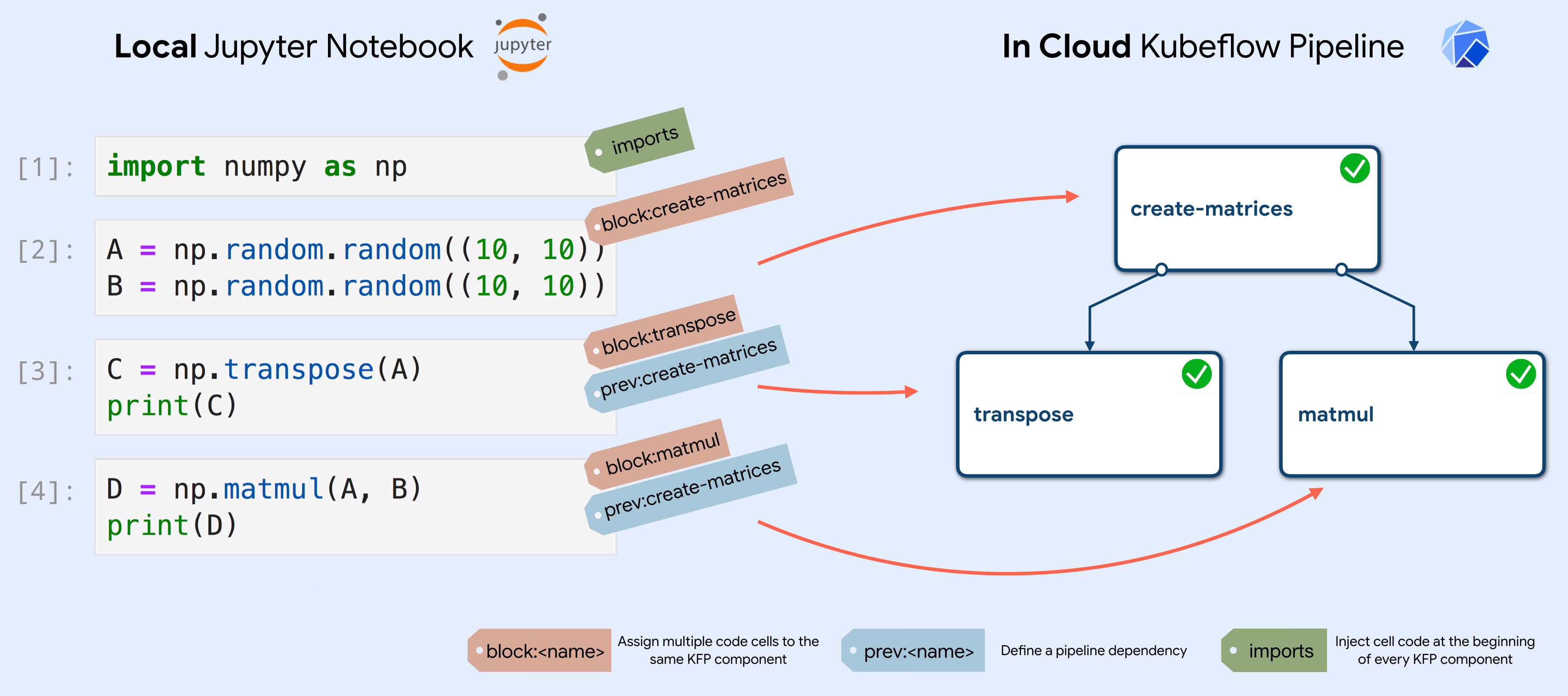
Kale takes as input the annotated Jupyter Notebook and generates a standalone Python script that defines the KFP pipeline using lightweight components, based on the Notebook and Cells annotations.
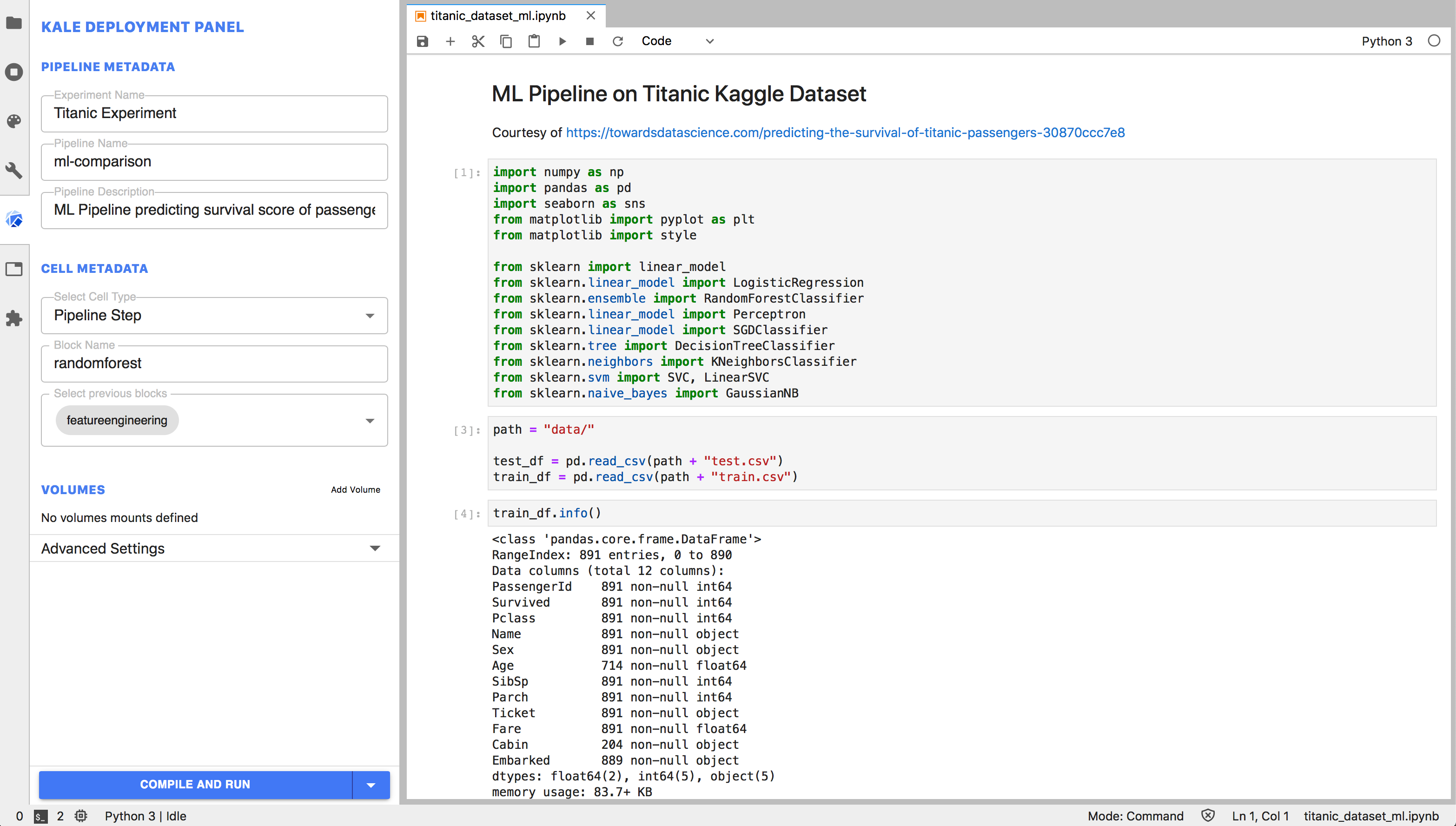
Annotating the Notebook becomes extremely easy with the Kale JupyterLab extension. The extension provides a convenient Kubeflow specific left panel, where the user can set pipeline metadata and assign the Notebook’s cells to specific pipeline steps, as well as defining dependencies and merge multiple cells together. In this way, a Data Scientist can go from prototyping on a local laptop to a Kubeflow Pipelines workflow without ever interacting with the command line or additional SDKs.
Data Passing
A question one might raise is: How does Kale manage to resolve the data dependencies, thus making the notebook variables available throughout the pipeline execution?
Kale runs a series of static analyses over the Notebook’s source Python code to detect where variables and objects are first declared and then used. In this way, Kale creates an internal graph representation describing the data dependencies between the pipeline steps. Using this knowledge, Kale injects code at the beginning and at the end of each component to marshal these objects into a shared PVC during execution. Both marshalling and provisioning of the shared PVC is completely transparent to the user.
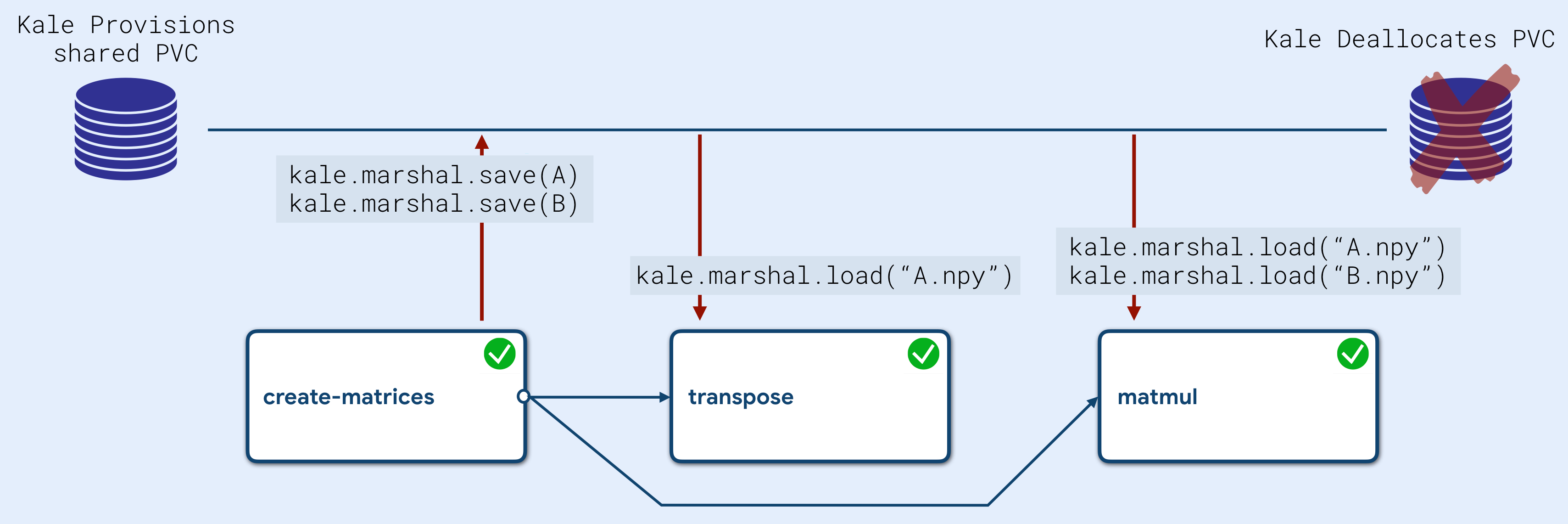
The marshalling module is very flexible in that it can despatch variable’s serialization to native functions at runtime, based on the object’s data type. This happens also for de-serialization, by reading the file’s extension. When the object type is not mapped to a native serialization function, Kale falls back to using the dill package, a performant general purpose serialization Python library (superset of pickle), guaranteeing high performance in terms of disk space and computation time.
Modularity and Flexibility
Kale’s output is a self-contained executable Python script, written with the KFP Python DSL to declare lightweight components that ultimately become the steps of the pipeline. All the required code generation is done via Jinja2 templates — generating Python with Python!
This solution makes Kale very flexible and easy to update to new SDK APIs and breaking changes. All the modules that read the annotated notebook, resolve data dependencies and build the execution graph are independent of KFP and Kubeflow in general. Ironically, Kale could be adapted pretty easily to any workflow runtime that makes available a Python SDK to define workflows in terms of Python functions.
Installation and Deployment
Q: How do I install Kale in my Kubeflow cluster / MiniKF?
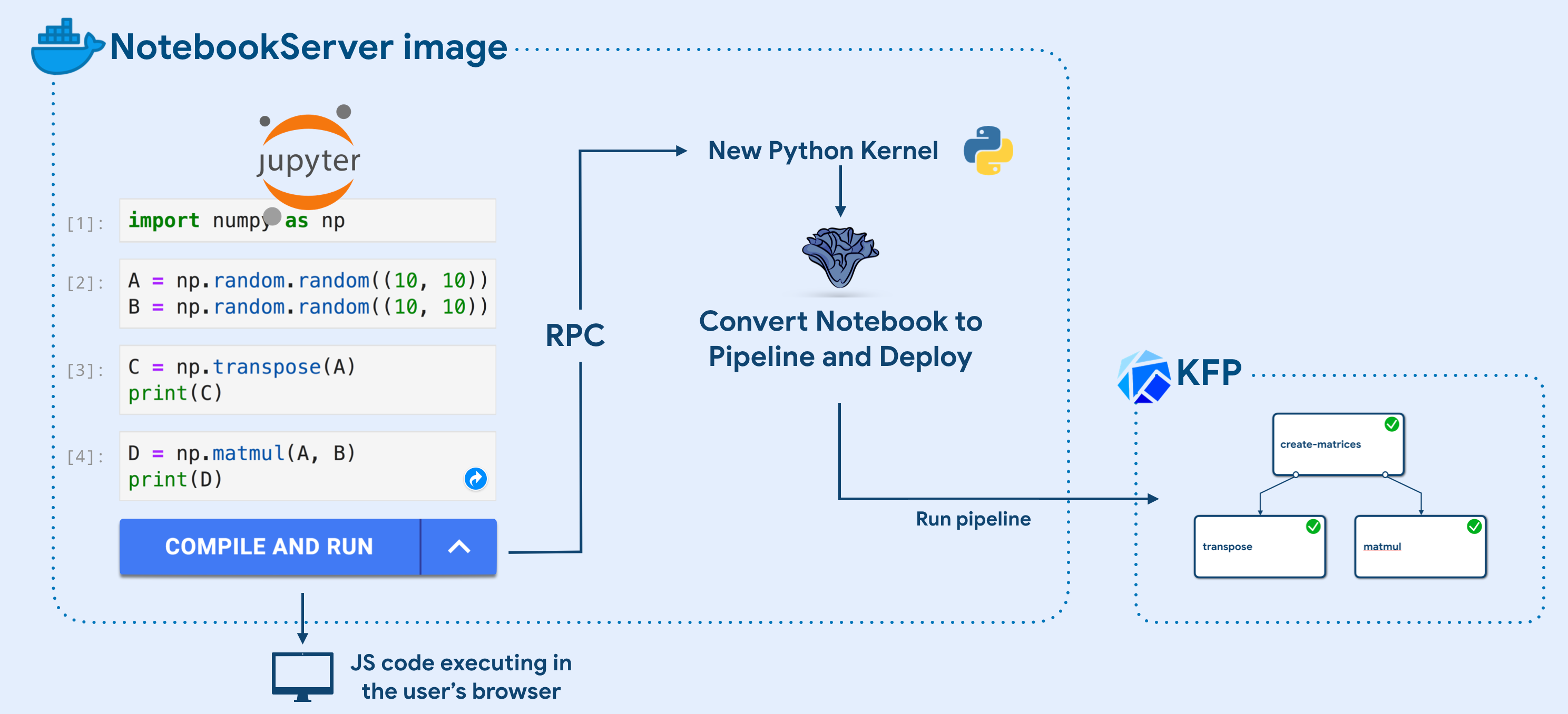
Kale does not require any cluster spec or new service to be installed. In fact, all you need to get started is a Docker image! Specifically, one just needs to take a NotebookServer image (used to spin up new Notebook Servers in Kubeflow) and extend it with Kale and Kale JupyterLab extension:
FROM <notebook-image>:<tag> # Install Kale python package globally RUN pip install kubeflow-kale # Install Kale JupyterLab extension RUN jupyter labextension install kubeflow-kale-launcher
The Jupyter extension is code that executes in the user’s browser, irrespectively of where the cluster is. For this reason, whenever an interaction with the cluster or the Kale Python backend is needed, we create a new Python Kernel in the background and execute an rpc method call to Kale, in order to execute some login inside the NotebookServer container.
Latest Developments
We have been working hard to make Kale the simplest way to approach Kubeflow Pipelines. Here is a (incomplete) list of the major new features introduced in the past few months:
1. Revamped metadata management
Kale now allows pipeline metadata to be stored and retrieved from Notebook metadata. Cell tagging language has been revised to be more flexible and expandable in the future.
2. Data Passing
Kale automatically provisions a new PV or uses an existing workspace volume to save marshal data.
3. Reusability
The codebase has been refactored to be used as an imported library. Each step in the conversion from Notebook to deployment can be programmatically called as an API.
4. Pipeline Parameters
Kale introduces a new tag parameters, used to transform code cells that contain just variables into reusable pipeline parameters.
5. Data Versioning and Snapshots
Integration with the Rok client when running Kale inside MiniKF. Kale can support these kind of workflows:
- Identify existing workspace/data volumes in the Notebook Server, snapshot them and mount them into the pipeline steps. In this way the Notebook workspace of the user (installed libraries, data, source files, …) are preserved in the running pipeline.
- Snapshot arbitrary volumes at the end of the pipeline run.
- Snapshot volumes at the beginning of each step run, providing a convenient way to recover the Notebook (data) state before the step failure for debugging.
6. JupyterLab extension
- Left panel to apply pipeline metadata (experiment, name, volumes) and run compilation, upload and run commands from the UI.
- The UI run Kale in the background by issuing Python commands in the running kernel, through a custom rpc module built into Kale.
- Cell Metadata UI control are placed on top of each Notebook Cell and color coded hints are provided to improve the readability of the KFP workflow on top of the Notebook.
- Upload, Run and Snapshot operations are run asynchronously and provide progress information to the user.
7. Packages
- Kale is packaged and uploaded to PyPi as kubeflow-kale
- Kale JupyterLab extension is packaged and uploaded to
npmas kubeflow-kale-launcher
Showcase
Kale is going to be showcased at KubeCon San Diego next week. Be sure to checkout the tutorial:
From Notebook to Kubeflow Pipelines — An End-to-End Data Science Workflow — on Thursday 21st.
We will showcase an end-to-end ML workflow on MiniKF, providing a seamless experience with the integration of Kale with Arrikto’s Rok data management platform for data versioning.
After the conference, we will update this post with a Codelab document link where you will be able to traceback all the content and reproduce the tutorial.
Post-conference update: Codelab link
For more information about the project, installation and usage documentation, head over to the Kale github organization.
For examples and use cases showcasing Kale in data science pipelines, head over to the Examples Repository.
Thanks to Ilias Katsakioris, Kostis Lolos and Tasos Alexiou (Arrikto) for their amazing contributions to the project, Chris Pavlou (Arrikto), Thea Lamkin and Jeremy Lewi (Google) for contributing to this post.
Deeply grateful to Valerio Maggio, Cesare Furlanello (MPBA — Fondazione Bruno Kessler) for helping in conceptualizing and fostering Kale.

